Making sense of drug combinations with mathematical models
EU-LIFE Science Newsletter 3/2017
News from the Institute for Molecular Medicine Finland (FIMM), Finland
Combinatorial therapies can tackle the drug resistance problem in cancer. Researchers at FIMM are developing novel informatics tools and pre-clinical testing platforms to facilitate the discovery of most promising targeted drug combinations among the exponentially increasing number of possibilities.
Drug resistance is a global problem in the treatment of many complex diseases including cancer. With heterogeneous genetic and epigenetic profiles, cancer can easily develop drug resistance by the activation of multiple compensating pathways as a result of homeostasis, or via the expansion of drug-resistant sub-clones. To reach long-term treatment efficacy, multi-targeted drug combinations are needed.
For pre-clinical evaluation of the combination effects, cancer cells are tested with multiple drugs in vitro or ex vivo. FIMM’s drug combination profiling platform currently enables the testing of more than 500 anticancer compounds including both FDA/EMA-approved drugs and investigational compounds. The platform is routinely being applied not only for cancer cell lines but also patient-derived samples.
FIMM Group Leader Jing Tang’s and Tero Aittokallio’s research teams work on the challenges brought by the increasing amount of drug combination screening data. They feel that the bottleneck has now moved to the informatics tools to visualize and analyze the data, and more critically, to facilitate the prioritization of synergistic drug combination hits for further validation.
Typical drug screening approach includes testing each pair of drugs at multiple concentrations, followed by a phenotypic assay of cell viability or toxicity. The resulting response profile captures the interaction patterns of the two drugs, often represented as a dose-response matrix. A drug combination is then classified as synergistic or antagonistic, depending on the deviation of the observed combination response from the expectation under a reference model.
The existing reference models were developed for low-throughput data, with oversimplified assumptions that are often incompatible with the dose-response matrix design. To address these limitations, FIMM researchers proposed a novel reference model, named zero interaction potency (ZIP), which captures the drug interaction relationships by comparing the change in the potency and shape of the dose-response curves between individual drugs and their combinations.
To provide a convenient and interactive way of exploring and making sense of drug combination data, the teams also developed an R-package and a web-application called SynergyFinder, now freely available for researchers (see image below). Dr. Jing Tang believes that these implementations will greatly benefit many drug combinations studies.
Original article: Ianevski, A., He, L., Aittokallio, T. and Tang, J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics. (In press)
Further reading:
- Tang, J., Wennerberg, K., Aittokallio, T. (2015) What is synergy? The Saariselkä agreement revisited. Front Pharmacol 6, 181
- Yadav, B., Wenerberg, K., Aittokallio, T. and Tang, J. (2015) Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput Struct Biotechnol J 13, 504-13
- He, L., Kulesskiy, E., Saarela, J., Turunen, L., Wennerberg, K., Aittokallio, T. and Tang, J. Methods for high-throughput drug combination screening and synergy scoring. Methods Mol Biol (In press).
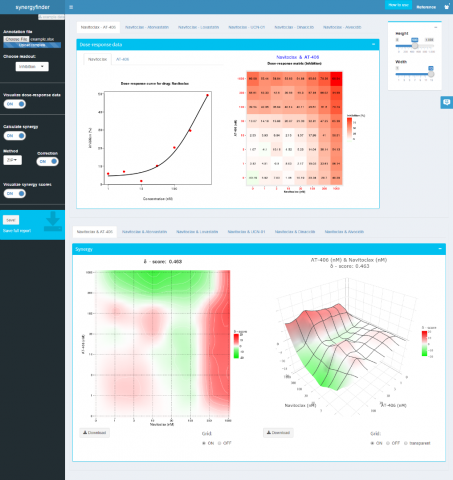
snapshot of SynergyFinder web-application (synergyfinder.fimm.fi)
